A rapid, reliable, and adaptable detection platform for COVID-19 and other pathogens using minimal reagents and preparatory steps
This detection platform uses double-stranded oligonucleotide probes to detect and quantify viral presence (e.g., COVID-19). In contrast to antibody tests, which rely on the host mounting an effective but delayed immune response to an invasive pathogen, Georgia Tech’s approach has the potential to identify infection during the earliest infection stages when an asymptomatic host can unknowingly spread the contagion. Unlike conventional flow cytometry samples, samples in this study did not require labeling of RNA targets or any wash steps (reagent use) to remove excess or unbound RNA. Thus, the overall simplicity of sample preparation and testing parameter requirements provides additional advantages to this rapid detection platform. Because this technology does not require high-demand sample collection supplies (e.g., swabs), shortages are much less likely to limit testing. Further, avoiding the use of reagents could reduce the likelihood of contamination seen in earlier tests.
- Earlier detection: Identifying the viral components themselves—rather than the body’s reaction to the viral presence (i.e., antibodies)—potentially improves early detection in asymptomatic subjects.
- Simpler process: Use of minimal reagents and fewer preparatory steps is designed to enable more efficient and facile sample preparation. Avoidance of “wash steps” aims to minimize target loss.
- Fewer disruptions: Avoiding the use of high-demand supplies (e.g., swabs) and reagents potentially reduces supply-chain and contamination issues.
- Fast results: Sample incubation of 15 minutes with an unlabeled target oligonucleotide sequence with sufficient complementarity would generate a fluorescent signal reporting a positive test.
Diagnostic markets for:
- Infectious diseases (e.g., COVID-19)
- Genetic disorders
- Microorganisms (e.g., bacteria, fungi, and viruses)
- Environmental (e.g., soil, water, etc.) DNA
Georgia Tech has developed an approach to potentially enable a robust, straightforward diagnostic platform with minimal reagents and preparatory steps. As society returns to various work environments and school campuses, the need for widespread COVID-19 testing continues to grow. Current testing platforms are susceptible to bottlenecks ranging from shortages in reagent supplies used in current molecular and polymerase chain reaction (PCR) and serological testing platforms to shortages in personnel trained and fully protected during test administration (e.g., collecting nasal swab samples).
Georgia Tech’s platform is intentionally simple in its handling and analysis, using high throughput equipment currently available in many hospitals and research settings (with more cost-effective desktop versions commercially available). Future implementation of these microsphere suspensions could allow for 1) scale-up to prepare testing kits for wide circulation among diverse communities, with each kit consisting of a small suspension volume (~10 µL; total of ~106 microspheres functionalized with double-stranded probes estimated to cost less than $0.25 in materials); and 2) expansion of sample source to saliva collected from the individual undergoing testing. Importantly, the platform is expected to be readily adaptable in its double-stranded probe sequence design to accommodate changes to detection platforms for future viral strains of COVID-19.
How It Works
The detection platform consists of colloidal particles functionalized with double-stranded oligonucleotide probes. Each double-stranded probe consists of a fluorescent dye-capped, immobilized sequence bound to a quencher-capped soluble hybridization partner. The proximity of the dye and quencher species inhibits or quenches fluorescence of the particle-immobilized sequence (i.e., initial “signal off” state). Upon incubation (less than 15 minutes), the quencher-capped sequence is competitively displaced by the unlabeled target. Once displacement is complete, the quencher-capped sequence is released into solution and thus allows fluorescence to occur (i.e., “signal on” detection of unlabeled target).
In contrast to popular PCR and conventional fluorescence-based oligonucleotide detection approaches, sample handling is simplified and streamlined by the following aspects of this double-stranded, probe-functionalized, colloidal particle-detection platform: 1) neither PCR nor 2) target sequence labeling are prerequisite sample-handling steps, and 3) no wash steps are needed once target sequence(s) are incubated with the colloidal suspensions. Finally, micron-sized colloidal particles are compatible with high throughput flow cytometry to enable fast detection (less than15 minutes) of the unlabeled target.
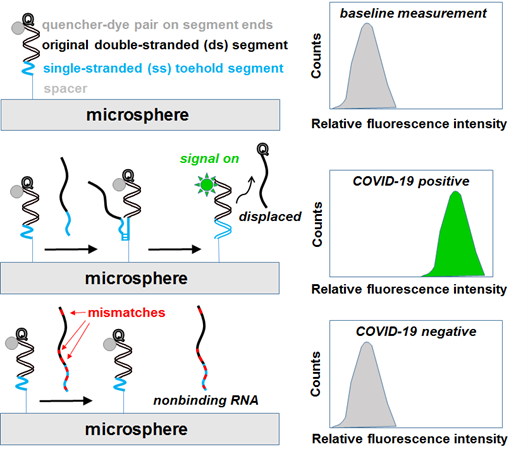
Schematics (left) and flow cytometry plots (right) of microsphere-immobilized double-stranded probes
