Georgia Tech inventors have developed an exhaustive means to search for possible on- and off-target cleavage sites for nucleotide-directed nucleases, such as CRISPR. CRISPR genome editing is currently being used across the globe in academic and corporate research labs facilitating basic research, making model organisms, and modifying plants and animals. Therapeutic use has been described for many disease indications, with work progressing through pre-clinical to the initiation of clinical trials. For each of these uses, the users are interested in maximizing the intended use and minimizing unintended editing effects, including off-target cleavage events. For all of these uses, it is critical that an exhaustive genome search is conducted to identify sites that may be cleaved by the nuclease or nucleases.
An exhaustive search for sites that may be cleaved must include both mismatches in sequence and the possibility of differences in length. These single or multiple difference in length between hybridization regions can be thought of or described as “bulges,” “indels” (insertions or deletions), “gaps,” or “skipped bases.” This technology allows the required inclusion of these searches along with searches for sites with mismatches. Numerous labs have verified that these sites can be cleaved and characterized these events (Lin, 2016).
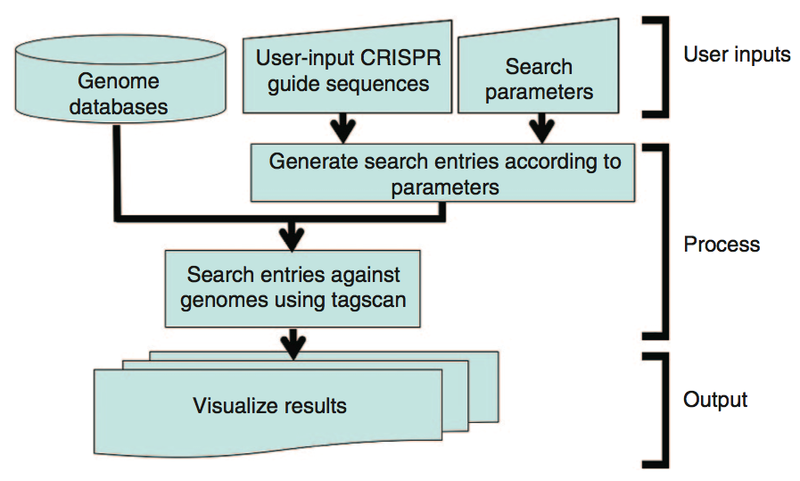
To facilitate these types of searches, Georgia Tech inventors also developed a method for identifying potential CRISPR-Cas9 off-target sites and facilitating their testing. The method enables searching genomes for potential off-target sites based on the user-supplied guide strand and input parameters. The method identifies potential off-target sites with mismatched bases and, unlike other methods, insertions or deletions when compared with the guide strand. The method aids in the identification and quantification of CRISPR-Cas induced off-target cleavage in cells. A search algorithm can scan a genome in seconds and provides web output in table format, with mismatches to the aligned query sequences marked in red. Links are provided to the University of California at Santa Cruz (UCSC) genome browser for each on-target and possible off-target locations to provide additional information on the site and flanking regions. In addition, all results are available for download to an Excel spreadsheet, allowing further analysis, sorting, and oligonucleotide ordering.
- Provides exhaustive genomic searches for off-target sites due to mismatches, deletions, and insertions
- Provides primers for experimental validation of predicted off-target sites
- Facilitates experimental testing using assays or deep sequencing of potential off-target sites given in the output
- Automatically designs primers to enable facile gel separation of the uncleaved and cleavage bands
- Includes the genomic reference sequence for comparison to sequencing results
- Facilitating and creating model systems for research and study of human disease and for sequencing potential off-target mutations
- Computational methods to identify, predict, and reduce off-target effects and unintended modifications at off-target sites during genome editing experiments
CRISPR-Cas systems are RNA-directed endonucleases for genome engineering, which are widely used for biological studies. CRISPR shows great promise for disease treatment with cellular correction of sicklecell anemia, cystic fibrosis, and other genetic defects. However, off-target effects can limit the usefulness of this technology. These off-target sites result from cleavage at similar genomic sites containing one or more base pair mismatches, insertions, or deletions. Identification of potential off-target sites is critical to optimize the CRISPR-Cas9 system and ensure that only intended sites are cleaved.