Researchers at Georgia Tech have developed a new algorithm for identifying and tracking cell membrane locations. This has the potential to automate electrophysiological measurements such as patch clamping, the “gold standard” of single-cell electrophysiology. The procedure leverages DIC microscopy to examine in vitro brain slices. Data is processed through a novel deconvolution algorithm formulated as a regularized least-squares optimization with the goal of providing automated visual tracking of a target cell’s membrane to guide automated electrophysiology systems (e.g., robotic patch clamping systems).
Importantly, this unique process can sift through high levels of organic tissue interference and can operate in environments with moving tissue. The algorithm filters out noise in the current image and then deconvolves and segments that image to determine cell edges based on the data from images earlier in the sequence. The final stage is a segmentation based on that deconvolution, which is performed with simple thresholding.
- Highly accurate: Overcomes the challenges of tissue interference and movement for precise membrane tracking that can be used for automation
- Instantaneous: Surpasses earlier attempts at automating this process by effectively identifying and tracking cell boundaries in real time
- High fidelity: Operates efficiently and precisely even in dynamic, high-interference environments
- Drug design and development
- Research tool
Manual patch clamping is widely considered to be the most effective means of taking electrophysiological measurements from a single cell. Its process, however, is challenging and laborious. The potential for automating in vitro brain slice patch clamping lies in the use of microscope imagery to visually guide the membrane tracking process. DIC microscopy is one of the best methods for examining samples that are otherwise transparent, but high levels of organic tissue interference complicate its application. This Georgia Tech method uses a novel cell segmentation and boundary tracking algorithm that could automate this process in in vitro brain slices.
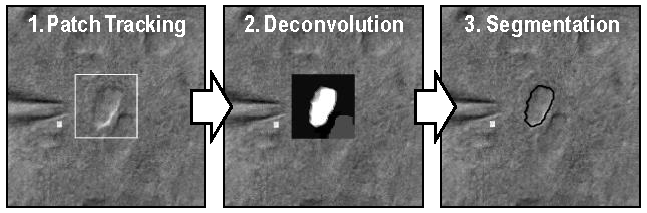
The three key phases of Georgia Tech’s novel method for tracking cell membranes using patch clamping and DIC microscopy.
