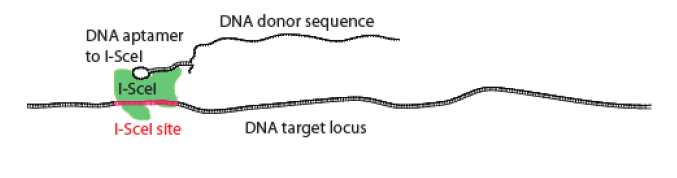
Georgia Tech inventors have developed a novel gene targeting system. Gene targeting typically is a technique to modify endogenous DNA by homologous recombination (HR). HR is typically inefficient in mammalian cell types; this ultimately limits the effectiveness of HR gene targeting. AGT, however, addresses the challenges associated with HR. AGT provides DNA aptamers that are capable of binding to a site-specific DNA binding protein and mediates the successful exchange of the genetic information between a donor molecule and a target locus.
The frequency of gene targeting was increased in this invention by attaching the targeting DNA to a protein involved in DSB or its repair. This concept was supported by the idea that by bringing the targeting DNA in close proximity of the DSB it would be preferentially used to repair the break, thus increasing the frequency. AGT incorporates a site-specific I-SceI endonuclease that generates a DSB, while repairing and binding to the target locus at the same time. AGT increases gene targeting for aptamer targets, DNA binding proteins, and proteins involved in recombination.
- Apatamers have unique secondary structure, allowing them to bind with high specificity and affinity to a specific target
- Utilizes intrinsic double stranded DNA break (DSB) and repair system, enabling targeted delivery of exogenous DNA in all eukaryotic cell types
- AGT development could lead to enhancement of gene editing technologies and ultimately disease-targeted therapy
- Functional analysis of genes
- Directed therapy for genetic disorders
Gene editing is a type of genetic engineering in which DNA is inserted, deleted, and/or replaced in the genome of an organism by a nuclease. There are currently four classes, including CRISPR-Cas9 system, Zinc finger nuclease (ZFN), transcription activator-like effector-based nucleases (TALEN), and meganucleases. AGT, also known as aptamer-guided targeting, should enhance all current classes of gene editing, increasing their effectiveness.